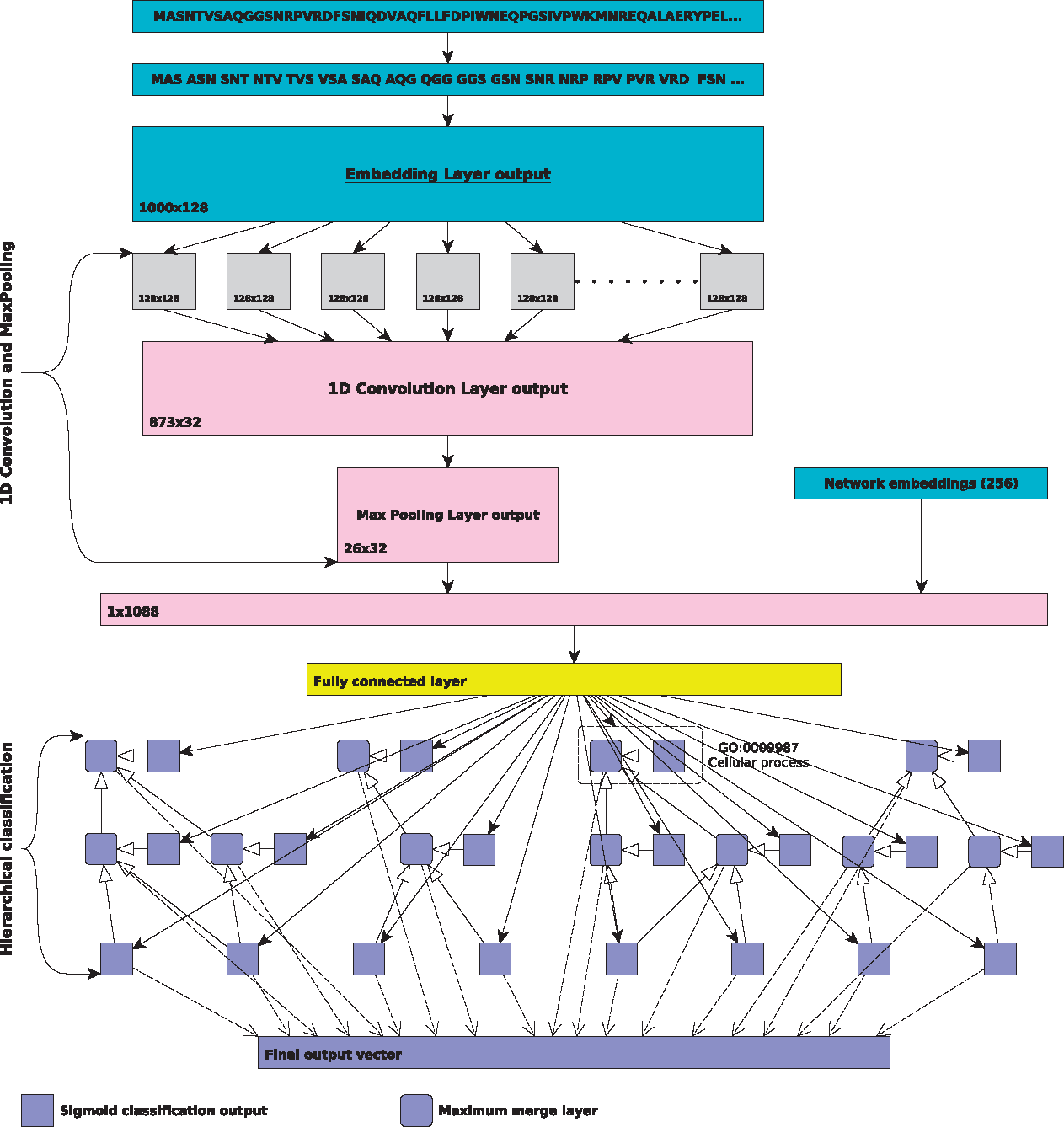
 DeepGO Neural Network Model
DeepGO Neural Network Model
DeepGO: predicting protein functions from sequence and interactions using a deep ontology-aware classifier
Abstract
Motivation A large number of protein sequences are becoming available through theapplication of novel high-throughput sequencingtechnologies. Experimental functionalcharacterization of these proteins is time-consumingand expensive, and is often only done rigorously forfew selected model organisms. Computational functionprediction approaches have been suggested to fillthis gap. The functions of proteins are classifiedusing the Gene Ontology (GO), which contains over40 000 classes. Additionally, proteins have multiplefunctions, making function prediction a large-scale,multi-class, multi-label problem.ResultsWe have developed a novel method to predict protein function fromsequence. We use deep learning to learn featuresfrom protein sequences as well as a cross-speciesprotein–protein interaction network. Our approachspecifically outputs information in the structure ofthe GO and utilizes the dependencies between GOclasses as background information to construct adeep learning model. We evaluate our method usingthe standards established by the ComputationalAssessment of Function Annotation (CAFA) anddemonstrate a significant improvement over baselinemethods such as BLAST, in particular for predictingcellular locations.
