 PhenoGO rule patterns
PhenoGO rule patterns
Abstract
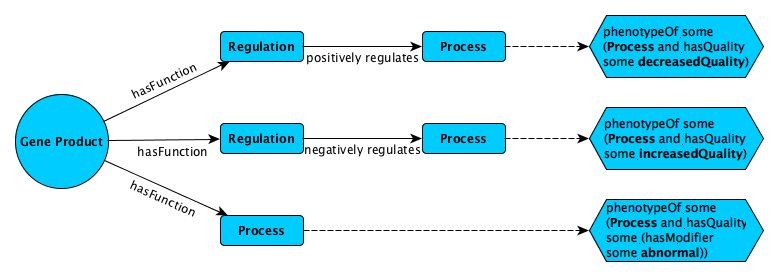
MotivationFunction annotations of gene products, and phenotype annotations ofgenotypes, provide valuable information aboutmolecular mechanisms that can be utilized bycomputational methods to identify functional andphenotypic relatedness, improve our understanding ofdisease and pathobiology, and lead to discovery ofdrug targets. Identifying functions and phenotypescommonly requires experiments which aretime-consuming and expensive to carry out; creatingthe annotations additionally requires a curator tomake an assertion based on reportedevidence. Support to validate the mutual consistencyof functional and phenotype annotations as well as acomputational method to predict phenotypes fromfunction annotations, would greatly improve theutility of function annotations.ResultsWe developed a novel ontology-based method to validate the mutualconsistency of function and phenotypeannotations. We apply our method to mouse and humanannotations, and identify several inconsistenciesthat can be resolved to improve overall annotationquality. We also apply our method to the rule-basedprediction of regulatory phenotypes from functionsand demonstrate that we can predict these phenotypeswith Fmax of up to 0.647.
